In Module 2 of 'Organising your DNA data and determining match groups' we discussed ways to organise data and a process for working with My Heritage to identify match groups. Before moving to Module 3, make sure you understand the theory of segment triangulation and have established a method for retaining records of your DNA research, to avoid rework and improve your productivity.
The purpose of undertaking these exercises is to consolidate your understanding of how to determine triangulated segments, identify potential match groups and whether a match is likely to be a false positive. By utilising the tools at My Heritage you can practice analysing your matches manually to ensure you appreciate the underlying theory. Remember, whilst shared matches are 'CLUES' of a shared ancestor, only shared and triangulated segments are 'EVIDENCE' of a common ancestor.
My Heritage is a DNA testing site that allows uploads from other testing companies. It's free to upload, but if you are not a My Heritage subscriber you will be required to pay a small 'one off' unlock fee to access DNA tools. My Heritage has two downloads that will be useful for chromosome analysis. These are the basic reports that you should start with:
* DNA Matches list
* DNA Matches shared segments list
My Heritage also has a clustering tool which can assist you to determine where to start your research, but this is best utilised once you understand more about triangulation. The clusters are an indication of matches in common, however understanding segment triangulation and how triangulated groups are formed will assist you to analyse the clusters more fully, helping to identify the key matches in the cluster more likely to share a common ancestor.
First steps
The following activities are suggested to help you apply 'Module 2' in practice. First you will need to download 2 reports from your DNA matches page. Click the 'three dots' icon and request the top two reports, the 'DNA matches list' and the 'Shared DNA segments report'. They will be emailed to you as .zip files. Download to your computer and open the zip file, creating 2 new .csv files. To open a zip file – right click on the zipped file – click on extract all – save the extracted files to a folder on your computer.
Exercise 1: Using the Broad Approach by Total cMs - My Heritage www.myheritage.com
- Utilising the 'DNA matches list' you downloaded from My Heritage for your DNA kit, sort the Total cMs shared column (largest to smallest).
- Make yourself familiar with the data contained in the downloaded spreadsheet.
- Add 3 additional columns for side (eg P, M, Both), MRCA (Most Recent Common Ancestor) and notes. Delete or Hide unnecessary columns for manageability (optional).
- Give your spreadsheet a working title, ie Total cMs Broad Approach - My Heritage.
- Analyse at least your top 10-20 matches - those with the highest total cMs (ideally >40cMs).
- Do you know if they are maternal or paternal (or both - ie share both sides, close relations) - mark your spreadsheet with the known side. Don't worry if you are not able to allocate sides at this stage.
- Think about the likely relationship, how many generations back in your tree might you expect to find the MRCA? (In Module 1 we discussed the Shared cMs tool to predict relationships? You might like to compare those estimates to those in the My Heritage prediction column).
- Notate your spreadsheet with the known or likely MRCA couple, if you can.
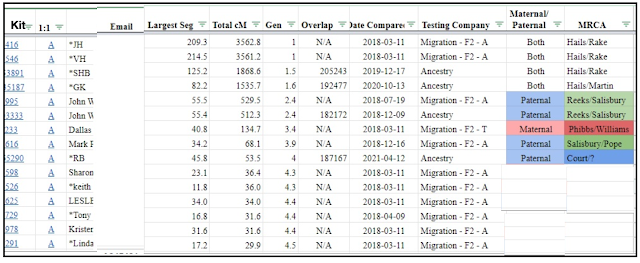
You may end up with a sheet that looks something like the one below - in this example I have highlighted my first unknown match in green. This match shares 60cMs, over 2 segments, the largest segment being 51.6cMs.
In Exercise 2 we want to examine an 'unknown' match who only shares on one segment (to make it easier to interrogate) so match #34 would be unsuitable. If your first unknown match also shares more than 1 segment, you will need to drill down further in the list to find another potentially suitable match to examine for this exercise.
Exercise 2: Using Chromosome Analysis (Specific Approach) - My Heritage www.myheritage.com
For this exercise we are going to utilise the 'Shared DNA segments report' and the My Heritage website.
Step 1. Prepare the spreadsheet:
- Open the 'Shared DNA segments report'.csv file you downloaded from My Heritage.
- Save it as a separate sheet and give it a working title, ie 'Chromosome Analysis - Specific Approach MH'. We will be amending this version, but we also want to retain the original 'Shared DNA segments report'.
- In your newly created 'Chromosome Analysis - Specific Approach MH' sheet, the columns we will be using will be Match Name, Chromosome, Start Location, End Location, Centimorgans.
- Delete or hide any unnecessary columns for manageability (optional).
- You may also wish to format the Centimorgans column to display the numbers showing one decimal point so that it is easier to read (ie 168 is the default number, re-format the column so it would appear as 168.0).
- Freeze the header row so that you can easily see and sort each column heading later.
- In your newly created 'Chromosome Analysis - Specific Approach MH' sheet, add columns for side (eg P, M, Both, I/F), TG (Triangulated Group), MRCA (Most Recent Common Ancestor) and notes.
- In your newly created 'Chromosome Analysis - Specific Approach MH' sheet, sort the centimorgans column into descending order to identify the matches with the largest segments, colour code any over 40cMs so you can identify them later (if you don't have any >40cMs just reduce your threshold);
- Go back to the original 'Shared DNA segments report' file from My Heritage. Examine the list and find the first 'unknown match' in the list who only shares on one segment (select one of your coloured matches if they fall into this category).
- In your 'Chromosome Analysis - Specific Approach MH' sheet, you can resort the matches by name to see how many chromosomes they match on. Otherwise you can always go back to the 'DNA matches list' (Exercise 1 by Total cMs) which lists the number of shared segments. (NOTE: My first unknown match highlighted in green above shares 2 segments so is not suitable for this exercise).
- In your 'Chromosome Analysis - Specific Approach MH' sheet, highlight your unknown match, in a different colour to the one you used for the cMs column.
- In your 'Chromosome Analysis - Specific Approach MH' sheet, resort the sheet by Chromosome and start and end locations so that they appear in order.
- Navigate back to your unknown match (Control/Command F - type in the name).
- See how many matches there are overlapping on the same segment shared by your unknown match.
- If there are a large number, double check that these are not in a known false positive region, or pile up areas. The chromosome map at DNA Painter is probably the easiest place to check this. If it is in a known false positive region, use a different match for the exercise, otherwise it may be too time consuming for what we are trying to demonstrate. Make a note in the notes column.
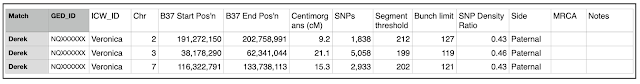
- Your sheet should look something like the one below.

Step 2. Identify the 'unknown match' you want to start with from Step 1 - Match A:
- Login to My Heritage site and navigate to your DNA Matches list.
- Find the 'unknown match' in your DNA Matches list on the MH site. We will call this match 'Match A'. Double check they only share on one segment and appear to have at least one triangulated match with other matches (look for the symbol in their shared match list).
- Decide on a name for the triangulated group you hope to identify for Match A from this exercise - it could be TG001-Side A (or whatever name you choose - it can reflect maternal/paternal if you know that information - eg M_001 or something similar).
- Go back to your 'Chromosome Analysis - Specific Approach MH' sheet that you created in Step 1.
- It should already be sorted by chromosome, then start and end locations.
- Find 'Match A' and put the TG number you created in the TG column.
- Identify the start and end locations of 'Match A's' shared segment area - eg Chromosome 18 from 11.4 - 59.0 and see how many matches have overlapping segments in this segment area. It may be helpful to colour code this group for easy identification of all the matches that you are now going to check at My Heritage to start the sorting process.

Return to your DNA Matches list on the My Heritage site. For the next part of the exercise you need to look for these two symbols on your My Heritage DNA match page, check where they appear before you start step 3.
Part 3. Identify likely triangulated groups from shared matches with triangulated segments.
- On the 'My Heritage DNA Matches' page, open up the comparison page between you and Match A, by clicking the purple 'Review DNA Match' button. Double check that you only share one segment with you selected match.
- As you look down the 'Shared Match List' on the My Heritage site, You and Match A will share at least one triangulated segment with anyone who has the TG symbol showing on the right. For this exercise - because we know Match A only shares one triangulated segment with you, all the matches with the triangulation symbol MUST BE matching on the same chromosome, so you and Match A and each match showing the symbol are forming a single triangulated group. You can click the TG segment symbol to check.
- We are now going to identify every shared match that has the triangulation symbol, this is evidence the 3 people have a triangulated segment (you, Match A and the match with the TG symbol). It is highly likely everyone who triangulates with you and Match A will also form a larger triangulated group, all being connected via a common ancestor.
- Review the list of shared matches looking for every shared match who has the 'triangulated segment symbol'.
- If you know a side or the expected line for the common ancestor, you may wish to add an appropriate coloured dot as you work through this exercise for the triangulated matches ONLY.
- For each match with the TG symbol, go back to your 'Chromosome Analysis - Specific Approach MH' sheet and notate the TG number in the TG column.
- Continue to work through the 'Shared Match List' on the My Heritage site, looking at all the shared matches between yourself and 'Match A' (keep pressing 'Show more DNA matches' when you get to the bottom of each page) and identifying those with the triangulated symbol and going back to the spreadsheet to notate the TG number the same as we did before. You can click on the TG symbol to see which segment is triangulating, but they all should be on the same chromosome and in a location common to Match A because we chose a match sharing only one segment. NOTE: Make sure you stay on the match page for 'Match A' and don't accidentally start reviewing shared matches of a different match by mistake.
- The triangulated symbol (shown in the shared match list) is telling you that you have at least one triangulated segment. Because we chose a match who only shared on one segment there will be no need to keep double checking the chromosome, as they should all be triangulating on the same segment. (This saves time and we will check the whole group later). For matches sharing more than one segment you will need to check the TG, this will be a much more time consuming exercise.
- As you work through the list, for those matches that have a tree, look to see if you can identify common names, locations or (if you are very lucky) the shared MRCA. Notate your master spreadsheet as you go. It is up to you whether you record these notes on the 'DNA matches list' or the 'Shared DNA segments spreadsheet', the MH site or all three!
- When you have exhausted all matches with the TG symbol, sort your spreadsheet by the TG column and review how many matches are noted as all being in the same triangulated group?
- Colouring code the names in the TG may assist in knowing you have reviewed them (optional);
- All members of this TG should share a common ancestor.
- After completing the rest of this exercise, come back to this group and look for common names or locations and try to find any genealogical connections within the group.
- Your 'Chromosome Analysis - Specific Approach MH' sheet should now look something like this.

NOTE: In some cases you might find that you have very large numbers of matches appearing in the overlapping segment area. If so, you can reduce the number of matches you compare in the second part of the exercise to a more manageable level by reducing the segment size to say >10cMs (this is a judgment call by you). If you do this, you will not be able to discard all false positive segments as you will not have done the complete comparison exercise.
Remember what we are trying to demonstrate for a valid triangulation is that A matches B, B matches C and C matches A, we need to do 3 comparisons. When we extend that out to a group, we need to do many more comparisons, ie for a 4th match, we need to ensure A matches B, A matches C, A matches D, B matches C, B matches D and C matches D, so there would be 6 comparisons. As the group gets larger the more comparisons we need to do to ensure every member is part of the group.
Part 4. Review the other side of the chromosome - starting with Match B (see below):
- After reviewing all the triangulated segments for 'Match A' at My Heritage, go back to your 'Chromosome Analysis - Specific Approach MH' and re-sort it by chromosome number and segment location.
- Look for the colour coded area indicating all matches that had overlapping segments with 'Match A'.
- Find the largest segment match in the list that has not been marked as belonging to the first TG group. We will call 'Match B'. If possible, find one that matches only on one segment to make the comparison process easier.
- If Match B matches on more than one segment, you will need to check each potentially triangulated segment and record only those on the opposing side of the same chromosome as Match A.
- Repeat the process in Part 3 for this match. Because 'Match B' did not triangulate in the first exercise, they must be a match on the other side of the chromosome, or are a false match. Make sure they triangulate with at least one other match. If they don't choose someone different.
- Decide on a name for this second triangulated group - it could be TG001-Side B (or whatever name you choose - it can reflect maternal/paternal if you know that information - eg P_001 or something similar).
- Notate each match with the TG number on your shared segment spreadsheet in the TG column.
- If you end up with smaller TG subgroups on the alternate side (different overlapping areas) you can number them B1 and B2.
- Colour code the names of the group in another colour so you can see who is included.
- Sort the 'Chromosome Analysis - Specific Approach MH' by the TG column and it should look something like this.

Part 5. Review what's left:
- Where there are TG's on both sides of the chromosome, any matches who do not match either side WITHIN THE SAME SEGMENT AREA can be marked as false positives, notate the side column F (False) or I (IBS/IBC).
- Remember in this example we chose a match who matched only on one segment so if they don't triangulate with either of the two groups they are a false match. That is not to say if they shared on multiple segments some of the other segments might be valid segments.
- In any remaining segment area where there is a TG on only one side of the chromosome, the opposing side should be left blank as we cannot determine anything about the validity of those segment matches at this stage. The alternative is to mark them to the opposing side, until further work is done, but you need to remember that many of them will end up being false positives.
- Your final list might look something like the one below.
- Retain the spreadsheet for future reference and build on it with further research.

In this example the segment area for Match A was 11.4 - 59, whilst Match B shared 9.7 - 24.2. We found two triangulated groups, one on either side of the chromosome. As we were able to identify the MRCA for some of the matches we were able to allocate them to maternal (TG_M_001) and paternal (TG_P_001) groups.
Because we did not investigate the second side of the chromosome between 24-59, this means all the matches who did not triangulate with TG_P_001 in this segment location area must be either maternal matches or potential false positives.
Part 6. Go back to the end of step three and explore your triangulated group for the MRCA (optional).
Is the whole group triangulated?
You can now go also back to the 'triangulated match tool' on the My Heritage site and check to see if all matches in the group you have identified (TG001-Side A) triangulate with each other. If the first segment was quite long, you may find the triangulations are in sub groups, like in the diagram below. You will need to play around with your match comparisons to identify the subgroups, but eventually you might see this sort of pattern.
This is demonstrating that whilst everyone is triangulating with 'Match A' (red), they are coming into the group at different levels. You and 'Match A' share the longest segment, so 'Match A' is probably a closer relation to you and your shared MRCA couple. The other matches may reflect matches to the same MRCA couple or perhaps a segment belonging to an older ancestor from one of the ancestors in that MRCA ancestral couple.
If you can identify the MRCA couple for Match A, others in each of sub groups could be segments coming from any of the 4 parents of that couple depending upon whether there was a recombination event in the segment area for Match A.
For example if Match A (red) was a 2nd cousin, they share your great grandparents. The subgroups could belong to either of those, or potentially different 2nd great grandparents depending on where recombination events occurred. Whilst I would initially call this one triangulated group for the entire length of Match A, if you identify the subgroups as belonging to more distant ancestors, you may wish to rename your subgroups as separate TG's.
Read more about reviewing your DNA matches at
My Heritage.
Next Steps - the quick way!
After you have mastered identifying your triangulated groups, access the 'My Heritage Auto Clusters' tool under the DNA tools menu. Remember that these are shared match clusters and need to be examined carefully to identify triangulated groups.
- Check to see if the match you selected for Exercise 2 appears in the cluster report;
- Who else is in the cluster?
- Which matches in the cluster are triangulated and who is only a shared match?
- Explore some new groups (optional);
- Don't forget to add any analysis to your master spreadsheet for future reference.
Need a bigger challenge?
Try this exercise from
Jim Bartlett - Triangulating your genome using My Heritage (Segment-ology, Dec 2020). I'd suggest attempting one chromosome to start, probably one of the smaller ones, Chromosome 20 might be good as it doesn't have any known false positive regions.
Veronica Williams
First Published: 21 Sep 2021
Last Updated: 19 December 2023