If you are like me, you have a couple of distant ancestors that are brick walls or where the paper evidence is not being confirmed by your DNA analysis. Is your paper evidence right, but there are just not enough descendants in the DNA mix? Or, is there a problem with your genealogy and your ancestor is someone else entirely?
How can chromosome analysis help you with this problem?
This blogpost is aimed at documenting the process to follow in these types of cases.
The plan and things to consider
- Where does the ancestor sit in your tree?
- What DNA test is most suitable, Y-DNA, mtDNA or autosomal DNA?
- How many descendants have tested?
- Have all these testers uploaded their data to a chromosome analysis site?
- Can you identify additional 'priority' testers that may assist in solving your problem?
- Are you confident that your DNA analysis to date (using the totals shared cMs approach), has confirmed your pedigree back to 2nd great grandparents for all lines? This ensures you have a solid pedigree base to start using chromosome analysis techniques.
- Have each of the generations between you and the subject ancestor been DNA confirmed?
- How many generations back on 'this line' do your confirmed DNA 'segments' go?
- Can you identify segment data for each generation?
Then take the following actions
- Identify triangulated groups in all of those segment areas;
- Interrogate each of the groups, searching for common names, locations, ethnicities, look for patterns between matches in each triangulated group;
- Build research trees, are there patterns or commonalities between triangulated groups?
- Ensure all 'DNA confirmed' segments can be 'walked back' on each chromosome, for each triangulated group;
- Continue to interrogate all triangulated groups with the aim to connecting them and revealing your shared ancestors.
Not all of their DNA got to you
We need to remember that due to the process of recombination, we don't all inherit DNA from our distant ancestors. Every generation loses 50% of our ancestors DNA. That's why it is so important to get as many of your relatives as possible to take a DNA test! Our genealogical tree is not the same as our genetic tree - we can still be genealogically related to other cousins but share no DNA. On average, you may have inherited about 3.1% of your 3rd great grandparents DNA - that is, if you got any at all!
How much DNA is available and who should I test?
DNA Painter has great tool called the Coverage Estimator, which can help to identify who is the best person to test to maximise the DNA for your 'target' ancestor. The following examples show the amount of DNA my family has for my great grandmother Abigail Courtney and her father, my 2nd great grandfather Arthur George Courtney.
Walking back the segments
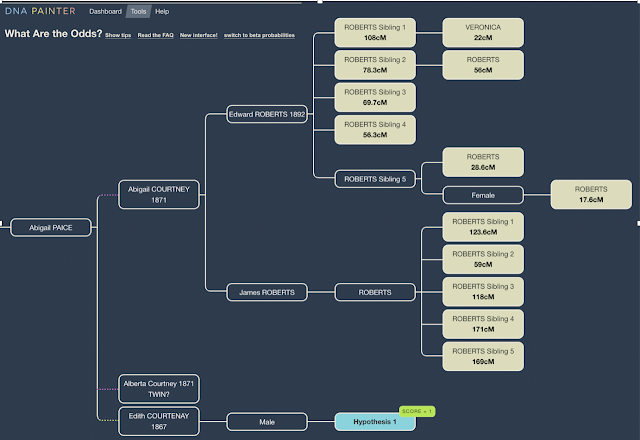
To start, you need a solid base of DNA confirmed matches that confirm your pedigree. 2nd cousins are ideal as they only share one grandparent with you, they need to have their results on a chromosome site such as GEDmatch, FamilyTreeDNA or My Heritage. Your goal is to identify the segments inherited from the 'target' ancestor. With matches at the 2nd cousin level you will be able to identify segments inherited from your shared great grandparents. In my case they are Edward Roberts and Abigail Courtney.
It is then a matter of separating the segments inherited from both ancestors to identify those that are on the 'target' ancestors line. By interrogating all triangulated segments for my Roberts-Courtney line (the line shared with my 2nd cousins) I am seeking to separate out Roberts line matches from Courtney matches. Then, we need to push each ancestors line back another generation so that we can isolate all triangulated segments belonging to the 'target' ancestor.
In my case, once I have identified segments inherited from Abigail Courtney, I then need to continue to interrogate those triangulated groups to seperate out the segments she inherited from her mother. By again separating these out, we should only be left with groups more likely to have been inherited from her father George Courtney (our target ancestor).
As you work through this process it is recommended to continually analyse both your triangulated groups and AncestryDNA clusters. Finding a 'bridge' match in a triangulated group and also in an AncestryDNA cluster may help lead to other connected matches that may be relevant to your search. Try to amass as many clues as possible to connect your triangulated group matches to find their common ancestor. Look for common connections within each group and between groups, the names may not be known to you but they could be clues pointing to your 'target' ancestor.
The follow chart seeks to document my progress in identifying the parents of my 2nd great grandfather Arthur 'George' Courtney. It can be a long term exercise to push segments back, but eventually you will find that the genetic clues will reveal common surnames, locations or ethnicities that may give clues to to help you progress your genealogical research.

Genetic research principles are very similar to traditional research, you start from yourself and move back one generation at a time. Just as in traditional research we seek to obtain birth and death certificates for our ancestors, rather than just checking the index; so do we seek triangulated segment data as evidence of shared ancestors, rather than just having matches in common. This becomes much more important for distant ancestors such as 2nd great grandparents and beyond, where shared cMs are more difficult in pinpointing a relationship given the plethora of possible relationships and the possibility of multiple shared ancestors.
Getting the segment data needed
Often the key matches that will help us solve our mystery are on AncestryDNA and there is no segment data available. We need to be creative in our requests when asking these cousins to transfer their results to a chromosome site so we can analyse the segment matches between us. Offering to help them with the analysis of their own kits can be a good way to get them engaged. Alternatively, think about setting up a group project at FamilyTreeDNA that might entice a broader range of testers that could assist in solving your mystery.
My Genetic Research
You can read more about my DNA analysis on my various blogs.
* My biggest research challenge is my maternal 2GGF Arthur George Courtney. He has a whole blog dedicated to him, read more on Finding George Courtney c1841. The Research Summary provides an overview of my genealogical and genetic research.
* In researching George, I managed to identify another maternal 2GGF: read my post - What is our family name? Roberts, Baker or Dye?
* Our Irish ancestors are always a challenge due to missing records, but DNA can often piece it together. My Britton family from Fermanagh are an example. I have much more to document about this family but these two posts may give you some ideas about what's involved. Meet Catherine 'Kitty' Britton, thanks to the X chromosome and Reuniting our Britton family, our unnamed Patriarch.
More information
For more information regarding chromosome analysis techniques, refer to the Table of Contents for this blog.
Veronica Williams
25 July 2023
 Whether your oldest BRITTON ancestor is a male or female, DNA testing can help to identify your ancestral Britton line. However, you may need to apply different DNA analysis techniques before you can confirm your Britton pedigree and to be confident of valid connections back to their original homeland.
Whether your oldest BRITTON ancestor is a male or female, DNA testing can help to identify your ancestral Britton line. However, you may need to apply different DNA analysis techniques before you can confirm your Britton pedigree and to be confident of valid connections back to their original homeland.